Volume 24 Number 4
The importance of anaerobic bacteria in non-healing wounds
Malik Asif Hussain, Irani Udeshika Rathnayake and Flavia Huygens
Keywords Chronic wounds, wound healing, anaerobes, bacterial diversity, Gram-positive anaerobic cocci.
Abstract
Chronic wounds, with their polymicrobial nature, put a significant burden on health budgets worldwide. Bacterial presence in the wound bed is associated with poor healing. The role of anaerobic bacteria in wound healing impairment has not been definitively established, mainly due to lack of reporting resulting from culture-based limitations. Advanced molecular methods are more reliable and the presence of anaerobic bacteria can be detected.
We have analysed 207 wound swab samples from 38 non-healing wound patients. Samples were collected at the Queensland University of Technology (QUT) wound clinic (Brisbane) over a period of 12 weeks. Next Generation Sequencing (NGS) technology was used for determining bacterial diversity and abundance in these wound samples. We found multiple types of bacteria residing on the non-healing wound bed. Bacterial diversity results are discussed in this paper with a focus on the importance of anaerobic bacteria. Overall, different patients’ wounds harbour diverse bacterial populations. Similarly, the presence of anaerobic bacteria in the wound bed was also detected. Molecular methods are reliable and useful, particularly for organisms requiring special conditions for growth such as anaerobes. It is clinically important to determine the major microbes present in the wound bed and their relationship with wound chronicity.
Introduction
A wound is formed due to damage to the skin and when the normal protective function of the skin is lost1. Bacteria have been reported to affect the healing process by invading the wound surface, damaging the tissue and prolonging the inflammatory response2-5. A wound which remains for more than six weeks6 or which does not progress to healing in four weeks7 is classified as a chronic wound. Bioburden is a broader term which includes total bacterial load and diversity as well as any specific virulent or resistant organism. Wound healing has been reported to be affected by bioburden8.
Various types of bacteria have been reported to be present in the wound bed. Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus (MRSA), is the most common bacterial species involved in skin infections1. Gjodsbol and colleagues have studied 50 patients who had chronic venous leg ulcers. Their results show that these wounds mostly harbour a polymicrobial flora, as indicated by the presence of more than one bacterial species in 94.4% of the samples9. James and colleagues reported the presence of Staphylococcus sp., Enterococcus sp., Pseudomonas sp., Proteus sp., Citrobacter sp. and Enterobacter sp. using culture-based methods, while molecular analysis detected additional species10. The most common bacteria reported in another study are; Staphylococcus sp., Pseudomonas sp., Peptoniphilus sp., Enterobacter sp., Stenotrophomonas sp., Finegoldia sp. and Serratia sp.11. Likewise, Streptococcus sp., Porphyromonas sp., Anaerococcus sp., Prevotella sp., Stenotrophomonas sp., Finegoldia sp., and Serratia sp. have been reported in other studies as well12,13.
Anaerobic bacteria have been reported to be involved in wound chronicity and biofilm production2,14. The presence of high percentages of anaerobic bacteria has been reported in pressure ulcers15. Similarly, anaerobic bacteria have been identified as a major contributor to wound bioburden16.
Gram-positive anaerobic cocci (GPAC) are an important group of anaerobic bacteria and are associated with 25–30% of clinical infections17. Anaerobic bacteria need absence or limited availability of oxygen. They find a perfect environment in deeper layers of biofilm as oxygen diffusion is blocked by biofilm layers18. Currently, the most clinically important genera of GPAC are Anaerococcus sp., Finegoldia sp., Parvimonas sp., Peptoniphilus sp. and Peptostreptococcus sp.17.
In general, GPAC infections are mostly controlled by beta-lactamase inhibitors, carbapenems, cephalosporins and chloramphenicol. In addition, variable levels of resistance have developed against antibiotics such as penicillins, clindamycin and metronidazole19. This variability in GPAC resistance and other anaerobic bacteria highlights the need to identify which anaerobes are mainly present in the wound bed and their impact on wound healing, so that wound infections can be promptly treated with appropriate antibiotics after identifying the relevant species/strains.
It is important clinically to determine which microbes are associated with chronic wounds and to detect their virulence determinants. There is a need to further increase our knowledge and understanding of the bacterial role in acute and chronic wounds14. Currently, there is no ‘gold standard’ for determining the correlation between bacterial load/bioburden and wound chronicity. It is, therefore, very important to develop more robust and accurate methods, such as molecular methods, to quantify bacterial load and diversity in chronic wounds. Once these methods have been developed and validated, wound treatment plans can be modified to improve healing outcomes20. This study aimed to identify bacterial diversity in non-healing wounds using advanced molecular techniques with its main focus on anaerobes.
Methods
Samples: Wound swab samples were collected at the Queensland University of Technology (QUT) wound clinic, with ethics approval for human research (QUT project approval number 1000001255). Swabs were collected using the Z-technique and were stored at –80°C until further use. Wound nursing clinicians, specialised in wound care, at the QUT wound clinic, were responsible for wound swab sample collection. A set protocol for collection of Z-swabs was followed by all clinicians involved in collection of the samples.
Wound swabs were collected from patients undergoing chronic wound treatment (QUT wound clinic) over a period of 12 weeks in 2011. Patients whose wounds did not heal by week 24 from their initial presentation at the wound clinic, were selected for this study and were categorised as “non-healers” while “healers” were excluded from the study. In total, 207 swabs from 38 non-healing wounds were used for this study. Out of these 38 patients, 19 had mixed ulcers, seven had arterial ulcers, seven had venous ulcers, two had pressure ulcers, two patients had undergone amputation surgery and one patient’s wound aetiology was unclear. These wounds were located at different areas of the lower extremities and all patients were receiving standard wound care at the clinic, including silver, hydro-fibre, hydrogel and zinc paste dressings. Prior to swab collection, wounds were washed with water. It was acknowledged that the use of antimicrobial dressings is likely to influence the microbial flora.
DNA extraction and Next Generation Sequencing (NGS): DNA extraction from swab samples was done following the Price et al.21 protocol with additional physical and enzymatic lysis steps. PCR amplification was performed using fusion primers, which were designed from the universal 16S rRNA (prokaryotic small subunit ribosomal RNA). Samples were subjected to NGS using the Ion Torrent PGM platform. Sequencing data was integrated using the Mothur software program (http://www.mothur.org/) and was further analysed using the Calypso software program. These experiments were performed at the Central Analytical Research Facility at QUT.
Results and discussion:
The results show varying trends of bacterial diversity among different patients (n=38). Overall, the bacterial diversity changes with or without a dominant genus or genera; however, there were patients whose wounds were dominated by particular genera from the earlier stages of sampling and remained prevalent throughout the sampling course. There were multiple types of bacteria detected in our patients. In total more than 50 genera have been detected and the dominant genera present in different patients were Staphylococcus sp., Bacteroides sp., Anaerococus sp., Peptoniphilus sp., Porphyromonas sp., Fusobacterium sp., Prevotella sp. and Finegoldia sp. Thus both facultative and strictly anaerobic bacterial genera have been detected in this study. This is consistent with research findings in other studies. Additionally the polymicrobial nature of chronic wounds is also validated by these results.
This paper focuses on the importance of anaerobes only. The overall results of bacterial diversity and abundance from all 38 non-healing patients are part of a separate investigation. Likewise, the importance of biofilm in wound healing has been reported and discussed in recent literature. Anaerobes are also involved in biofilm production. Similarly, staphylococci and Pseudomonas species are also biofilm producers. Our research group has submitted an article reporting the presence and absence of common biofilm controlling genes, which includes a discussion about the role of biofilm in wound chronicity. The following figures show results of bacterial diversity in the form of bar charts and bubble plots using the Calypso software for patient #4001 and patient #4059 as representative samples. These two patients are presented as an example.
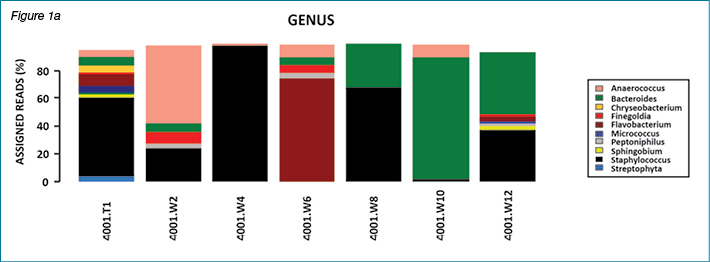
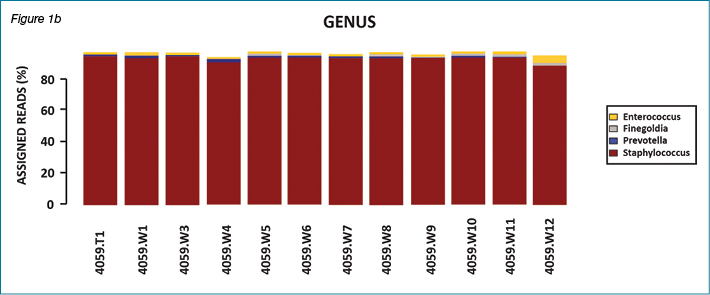
In patient #4001, multiple genera were present with variations over the course of the study while in patient #4059 there was dominance of one genus (Staphylococcus sp.) throughout the study period. This variation in type and dominance was observed for several patients, with the majority of patients presenting with changing bacterial flora over 12 weeks of sample collection.
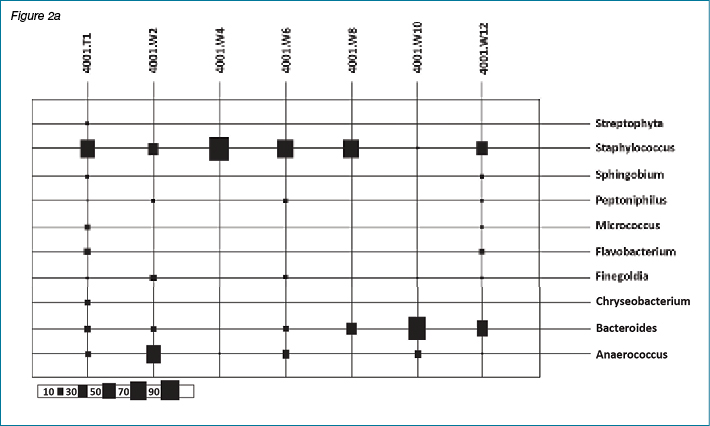
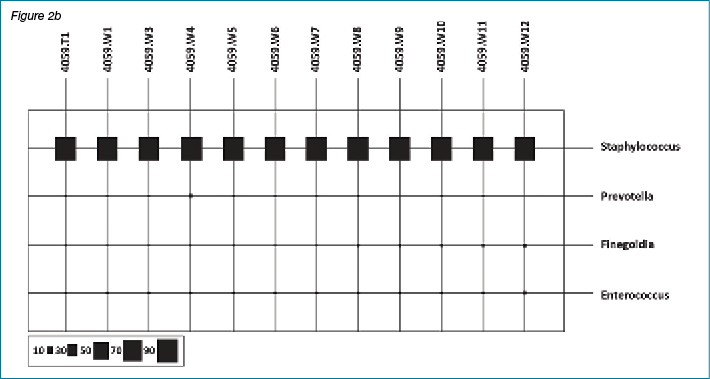
Figures 2a and 2b show results of bacterial diversity in the form of bubble plots, using the Calypso software, for patient #4001 and patient #4059. It is clear from these figures that patient #4001 has variation in bacterial diversity at different weeks with Staphylococcus sp. and Anaerococcus sp. the dominant bacterial genera present. On the other hand, patient #4059 clearly shows Staphylococcus sp. dominance throughout the course of the study. The scale under the bubble plot represents the percentage of each genus.
Culture-based techniques have been widely used to study wound bioburden and were regarded as the gold standard method for this purpose but molecular methods have produced more promising results17,22. Culturing is a slow and selective process that misses the majority of organisms including anaerobes, which are very important from the wound healing point of view11,23,24. Molecular diagnostic techniques provide a more diverse picture of the microbial content of wounds and/or biofilms11. Frank et al. proposed that it is worth combining culturing with molecular methods to achieve the best results22.
Advanced techniques, such as NGS, are able to reveal further details regarding the microbial flora of clinical samples, including that of chronic wounds11. Han and colleagues have compared culture-based methods and NGS, and they have reported the presence of three main bacterial species (on average) using culture-based methods while pyrosequencing on average detected 17 different genera. Most of the genera detected by NGS were anaerobic bacteria such as Anaerococcus sp., Finegoldia sp. and Peptinophilus sp.16.
Dowd and colleagues10 used pyrosequencing, shotgun Sanger sequencing and denaturing gradient gel electrophoresis for the investigation of bacterial diversity in chronic wounds and Gardner and colleagues12 used 16S rRNA sequencing for bacterial diversity studies13,15. The development of molecular methods such as PCR, multiplex PCR, 16S rRNA sequencing and NGS has improved the detection and identification of numerous organisms, including anaerobic bacteria17. As mentioned previously, we also detected the presence of more than 50 genera in total in all of our patients (n=38) with dominance of Staphylococcus sp., Bacteroides sp., Anaerococus sp., Peptoniphilus sp., Prevotella sp. and Finegoldia sp.
Anaerobic bacteria are usually cultured using blood or fastidious anaerobic agar and incubation periods vary from 48 hours up to 7 days17. Morphology, Gram-staining and sensitivity to antibiotics such as metronidazole are used to identify anaerobes in many laboratories19. Some GPAC species have been reported to have resistance to metronidazole, thus these organisms would be ignored if metronidazole sensitivity is a criterion for identification19. We are reporting the presence of anaerobic bacteria in our non-healing wound patient samples and suggesting their association with wound chronicity. The exact impact of anaerobes on wound healing as well as virulent determinants related to wounds requires further investigation. Wound management and treatment will improve by revealing details regarding wound microbial flora17.
The clinical significance of GPAC has been underestimated for years, mainly due to two factors. Firstly, culture-based techniques usually fail to report anaerobic bacteria due to their slow growth and specific growth requirements in the laboratory. Secondly, they are usually present in infections involving multiple bacteria, thus other known pathogens have been given more clinical importance and the presence of anaerobic bacteria has been overlooked17. Samples for identification of anaerobic bacteria require appropriate collection, transportation and strict anaerobic culture conditions17. Culture-based methods have limitations, therefore molecular methods should be developed25,26. Advanced molecular techniques have increased GPAC detection and established their involvement in clinical infections. Furthermore, molecular methods have detected many new species in this group resulting in changes in taxonomy and nomenclature17. The availability and cost related to molecular methods is an important issue. However, we recommend the development of a standardised protocol for the evaluation of non-healing chronic wounds.
Clinical relevance of findings
Based on the experimental results as well as the literature review, we are proposing the following:
- Molecular techniques should be preferred over cultural methods to obtain a better picture of bacterial diversity in the wound bed. This is particularly important for the identification of bacteria that require special transport and culture conditions such as anaerobic bacteria.
- Anaerobic bacterial infections need to be treated appropriately as they have been reported to delay healing and are involved in biofilm formation2,14. Furthermore, resistance to antibiotics has been reported in anaerobic bacteria27,28.
- We recommend regular monitoring of wound bed flora, particularly in non-healing wounds as our results show new bacterial types can grow over the course of treatment and replace the originally detected bacterial type(s). This means there might be a need to further add antibiotics or change them. Regular evaluation and change in the treatment plan has been found to reduce the total cost of treatment29. For this purpose, a clinical evaluation of wounds, involving wound treating physicians and staff is required. Clinical signs of infection such as redness and heat cannot be totally relied on as these are usually not very prominent in chronic wounds due to several factors, including diseases such as diabetes mellitus, and the use of drugs which suppress pain and inflammation30.
Author disclosure and ghostwriting
No conflicts of interest exist. The content of this article was expressly written by the authors listed. No ghostwriters were used to write this article.
Acknowledgements
We acknowledge the support of the Australian Government’s Cooperative Research Centres Program and QUT for this research.
Author(s)
Malik Asif Hussain*
MBBS, MAppSc (Research)
PhD(C), School of Biomedical Sciences, Faculty of Health, Queensland University of Technology
Brisbane, Qld, Australia
Institute of Health and Biomedical Innovation,
60 Musk Avenue, Kelvin Grove, Qld 4059, Australia
Email: hussain_amc2010@yahoo.com;
m1.hussain@qut.edu.au
Tel +61 490 151 350
Irani Udeshika Rathnayake
PhD
Research Fellow, School of Biomedical Sciences, Faculty of Health
Queensland University of Technology
Brisbane, Qld, Australia
Flavia Huygens
PhD
Associate Professor, School of Biomedical Sciences, Faculty of Health
Queensland University of Technology
Brisbane, Qld, Australia
* Corresponding author
References
- Percival SL, Emanuel C, Cutting KF, Williams DW. Microbiology of the skin and the role of biofilms in infection. Int Wound J 2012;9(1):14–32.
- Scales BS, Huffnagle GB. The microbiome in wound repair and tissue fibrosis. J Pathol 2013;229(2):323–31.
- Bhattacharya R, Xu F, Dong G et al. Effect of Bacteria on the Wound Healing Behavior of Oral Epithelial Cells. PloS One 2014;9(2):e89475.
- Ngo QD, Vickery K, Deva AK. The effect of topical negative pressure on wound biofilms using an in vitro wound model. Wound Repair Regen 2012;20(1):83–90.
- Murray RZ, Röhl J, Zaharia A, Rudolph M. The role of inflammation in cutaneous repair. Wound Practice & Research 2015;23(1).
- Frankel YM, Melendez JH, Wang NY, Price LB, Zenilman JM, Lazarus GS. Defining wound microbial flora: molecular microbiology opening new horizons. Arch Dermatol 2009;145(10):1193–5.
- McCarty SM, Cochrane CA, Clegg PD, Percival SL. The role of endogenous and exogenous enzymes in chronic wounds: a focus on the implications of aberrant levels of both host and bacterial proteases in wound healing. Wound Repair Regen 2012;20(2):125–36.
- Gardner SE, Frantz RA. Wound bioburden and infection-related complications in diabetic foot ulcers. Biol Res Nurs 2008;10(1):44–53.
- Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, Krogfelt KA. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 2006;3(3):225–31.
- James GA, Swogger E, Wolcott R et al. Biofilms in chronic wounds. Wound Repair Regen 2008;16(1):37–44.
- Dowd SE, Sun Y, Secor PR et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 2008;8:43.
- Rhoads DD, Cox SB, Rees EJ, Sun Y, Wolcott RD. Clinical identification of bacteria in human chronic wound infections: culturing vs. 16S ribosomal DNA sequencing. BMC infect Dis 2012;12:321.
- Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 2013;62(3):923–30.
- Wolcott R, Dowd S. The role of biofilms: are we hitting the right target? Plast Reconstr Surg 2011;127(Suppl 1):28S–35S.
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 2008;3(10):e3326.
- Han A, Zenilman JM, Melendez JH et al. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen 2011;19(5):532–41.
- Murphy EC, Frick IM. Gram-positive anaerobic cocci — commensals and opportunistic pathogens. FEMS Microbiol Rev 2013;37(4):520–53.
- Rasmussen K, Lewandowski Z. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol Bioeng 1998;59(3):302–9.
- Hecht DW. Anaerobes: antibiotic resistance, clinical significance, and the role of susceptibility testing. Anaerobe 2006;12(3):115–21.
- Grice EA, Segre JA. Interaction of the microbiome with the innate immune response in chronic wounds. Advances in experimental medicine and biology 2012;946:55–68.
- Price LB, Liu CM, Frankel YM et al. Macroscale spatial variation in chronic wound microbiota: a cross-sectional study. Wound Repair Regen 2011;19(1):80–8.
- Frank DN, Wysocki A, Specht-Glick DD et al. Microbial diversity in chronic open wounds. Wound Repair Regen 2009;17(2):163–72.
- Veeh RH, Shirtliff ME, Petik JR et al. Detection of Staphylococcus aureus biofilm on tampons and menses components. J infect Dis 2003;188(4):519–30.
- Landis SJ. Chronic wound infection and antimicrobial use. Adv Skin Wound Care 2008;21(11):531–40; quiz 41–2.
- Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001;14(2):244–69.
- Davies CE, Wilson MJ, Hill KE et al. Use of molecular techniques to study microbial diversity in the skin: chronic wounds reevaluated. Wound Repair Regen 2001;9(5):332–40.
- Sedano Gomez GE, Perez de Llano LA, Pita Carretero J. Necrotising pneumonia due to Finegoldia magna. Arch Bronconeumol 2011;47(1):54–5.
- Rosenthal ME, Rojtman AD, Frank E. Finegoldia magna (formerly Peptostreptococcus magnus): an overlooked etiology for toxic shock syndrome? Med Hypotheses 2012;79(2):138–40.
- Tan B, WX TE, Chong S, Chang Y, Song C, Lee V. An economic evaluation of chronic wound management in a tertiary hospital. Wound Practice & Research 2016;24(3):130.
- Siddiqui AR, Bernstein JM. Chronic wound infection: facts and controversies. Clin Dermatol 2010;28(5):519–26.



