Volume 5 Issue 2
The development of a real-time quantitative polymerase chain reaction (qPCR) method for the detection of Staphylococcus aureus in peripherally inserted central catheter (PICC) colonisation
Maddie Higgins, Jeremy Brownlie Li Zhang and Rebecca Ford
Keywords Central line-associated blood stream infection, CLABSI, peripherally inserted central catheter, PICC, nuc gene, Staphylococcus aureus, S. aureus, quantitative real-time polymerase chain reaction, qPCR.
For referencing Higgins M et al. The development of a real-time quantitative polymerase chain reaction (qPCR) method for the detection of Staphylococcus aureus peripherally inserted central catheter (PICC) colonisation. Vascular Access 2019; 5(2):23-28.
DOI https://doi.org/10.33235/va.5.2.23-28
Abstract
Background Peripherally inserted central catheters (PICCs) are susceptible to Staphylococcus aureus (S. aureus) colonisation and subsequent dissemination into the bloodstream, leading to central line-associated bloodstream infections (CLABSI). Current detection for S. aureus PICC colonisation relies on the use of traditional culture-dependent methods, including the semi-quantitative roll-plate culture method. However, the minimum time to detection is between 24–48 hours. Furthermore, a definitive diagnosis may take up to 7 days and is therefore not useful in guiding appropriate and timely patient management. A quantitative real-time polymerase chain reaction (qPCR) assay has the potential to overcome these limitations.
Methods A qPCR assay, targeting the nuclease (nuc) gene, was developed to detect S. aureus PICC colonisation. The sensitivity threshold of the assay was determined using purified S. aureus genomic DNA (gDNA) and validated using 41 clinical PICC samples which were compared to results from the roll-plate culture method.
Results The sensitivity threshold of the qPCR assay was 102 CFU/mL-1. From a total of 41 clinical PICC samples, S. aureus colonisation was detected from one PICC by both qPCR (103 CFU/mL-1) and the roll-plate culture method (103 CFU/mL-1). The qPCR assay processing time was less than 2 hours after bacterial gDNA isolation compared with 24–48 hours for the roll-plate culture method.
Conclusion This developed qPCR assay is an accurate and rapid method to detect S. aureus PICC colonisation. With further research, this method has the potential to be used in a clinical setting.
INTRODUCTION
Peripherally inserted central catheters (PICCs) are increasingly used for intermediate and long-term access in patients for the delivery of vital fluids, including blood products, medication and nutrition1,2. However, PICCs are susceptible to bacterial colonisation and subsequent dissemination into the bloodstream, leading to central line-associated bloodstream infections (CLABSI) which can occur with prolonged insertion time, patient immunosuppression, and the administration of parenteral nutrition3. Furthermore, microbial attachment on PICCs may occur within 24 hours of insertion, with microbial biofilm formation evident within 48–72 hours4.
A well-recognised complication of PICC usage, CLABSI results in increased morbidity, mortality, and the length and cost of hospital stays5-7. An estimated 250,000 bloodstream infections occur annually, with 80,000 CLABSI occurring in intensive care units each year8. These infections contribute towards mortality rates ranging between 12–25% in critically ill patients, with an estimated cost between $4,000–56,000 per episode9-11. Common causal pathogens of PICC colonisation include Staphylococcus aureus (S. aureus), Enterococci and Candida spp.12. In Australia, an estimated 7,000 S. aureus bloodstream infections occur annually, and are largely associated with CLABSI13. The infection is difficult to treat, may require removal of the colonised PICC, and often involves prolonged antibiotic therapy8. Furthermore, S. aureus is widely known for its disease-causing potential, including vascular, cardiac and pulmonary complications14, and is associated with 25–35% mortality despite strict efforts to minimise infection15. Consequently, there is a need for an accurate and rapid method to detect S. aureus PICC colonisation to guide appropriate and timely patient management and to reduce the potential for CLABSI16.
Routine detection of S. aureus PICC colonisation heavily relies on the use of traditional culture-dependent methods, including the semi-quantitative roll-plate culture method, employing selective media for direct enumeration or recovery of isolates after enrichment in selective broth17. A count of 15 colony-forming units (CFU) or more per plate indicates PICC colonisation, while a definitive diagnosis of CLABSI requires that the same organism, associated with PICC colonisation, grows from at least one percutaneous blood culture7,8. However, the semi-quantitative roll-plate culture method has several disadvantages. First, the minimum time to detection and identification of the causal organism is between 24–48 hours. Second, the sensitivity of this phenotypic method is limited to approximately 70% and even lower for fastidious and slow-growing bacteria18. Third, prior administration of antibiotic therapy may significantly impact the sensitivity of cultures19-21. These disadvantages can be overcome with the use of quantitative real-time polymerase chain reaction (qPCR), a time- and cost-effective high-throughput method that can be performed directly on isolated genomic DNA (gDNA) with a processing time of less than 2 hours22.
Previously, rapid, sensitive and specific qPCR assays for the detection of S. aureus have been developed targeting the nuclease (nuc) gene23. The main reason for targeting the nuc gene is that it is well conserved in S. aureus at the nucleotide level and hence evolutionary stable24. Studies have used the nuc gene to accurately detect S. aureus from blood cultures25,26, wound swabs, bodily fluids, tissue samples, urine, vascular access sites, and sputum27. In particular, the nuc gene has been used to successfully detect S. aureus gDNA in blood to diagnose bacteraemia22. However, this sequence has not previously been used to detect S. aureus colonisation directly from PICCs, after being withdrawn from patients, using qPCR.
There is a significant requirement for an accurate and rapid method to detect S. aureus PICC colonisation, allowing for more informed decision-making, with the potential for early intervention in the diagnosis of S. aureus CLABSI3. A qPCR assay using primers specific to the nuc gene has the potential to meet these requirements. Therefore, the aim of this study was to assess and validate a qPCR assay using primers specific to the nuc gene to detect S. aureus PICC colonisation. This was achieved by:
a) Assessing existing published literature to identify an appropriate target gene sequence, and to confirm the target amplification size of the gene sequence using conventional PCR;
b) Converting the target sequence into a qPCR assay;
c) Determining the sensitivity of the optimised qPCR assay to detect S. aureus in an environment that simulates a clinical setting; and
d) Validating the sensitivity of the qPCR assay by determining the detection level of S. aureus on PICCs derived from clinical samples.
MATERIALS AND METHODS
Clinical PICC samples were supplied from 41 in-patients at the Queensland Children’s Hospital, Queensland, including traditional polyurethane PICCs (Turbo-Ject Power-Injectable PICCs [Cook Medical; Bloomington, IN]) and hydrophobic PICCs (Bioflo® with Endexo® PICCs [Angiodynamics Inc; Queensbury, NY]). Ethical approval for the study was granted by Queensland Health Human Research Ethics Committee (HREC/15/QRCH/164) and Griffith University Human Research Ethics Committee (HREC/2016/077), and the trial was registered with the Australian New Zealand Clinical Trials Registry (ACTRN12615001290583).
Study inclusion criteria
Patients were eligible to participate in the study if they were less than 18 years of age, required a PICC inserted for treatment for more than 24 hours, and did not have a bloodstream infection at time of recruitment. We needed to have written informed consent from legal guardians and to be able to safely collect PICC materials at the time of the PICC removal. All aspects of PICC insertion, use, management and removal were completed by clinical staff in accordance with local clinical practice guidelines, including the use of 2% chlorhexidine gluconate in 70% ethanol for skin antisepsis28.
Sample collection
All PICC samples were collected under aseptic conditions by a qualified research nurse with experience in preparation of specimens for culture. The distal 2–3cm of the PICC was collected using sterile scissors, deposited in a sterile container, and immediately transported to the laboratory for culture using the semi-quantitative roll-plate culture method29. Descriptive information – including patient demographics, PICC utilisation, clinical characteristics and PICC complications – were collected from patients by a research nurse and stored on a secure online database30.
Identification of S.aureus nuc gene
Existing published literature was reviewed to identify the 16S nuc gene primers F (5’-GCGATTGATGGTGATACGGTT-3’) and R (5’-AGCCAAGCCTTGACGAACTAAAGC-3’) used in this study. The primers consisted of 21 and 24 bases, respectively, and were located within the 270-bp nuc gene, encoding the nuc gene. The nucleotide positions were 48–70 and 303–328, respectively24.
S.aureus AU-19 reference strain
One clinical isolate Staphylococcus spp. (S. aureus AU-19), previously sequenced using 16s rRNA analysis, was included in this study as a reference strain [Li Zhang, 2011, Nathan Campus, Griffith University] to evaluate the diagnostic performance of the qPCR assay.
S.aureus AU-19 gDNA extraction
A single colony of purified S. aureus AU-19 was suspended in 10mL of Luria broth and incubated overnight at 37˚C in preparation for gDNA extraction. Bacterial gDNA was extracted using the PowerSoil DNA Isolation Kit [Mo Bio; California, USA].
Conventional PCR: amplification of the nuc gene
Conventional PCR was used to confirm the target amplification of the expected size and sequence of the nuc gene with S. aureus AU-19 using the MyCyclerTM Thermal Cycler System [BioRad; California, USA]. All reactions were performed in triplicate and included both positive and negative controls. The PCR amplification was performed as per the manufacturer’s instructions31, using primers specific to the nuc gene24, and the PCR product was visualised using 2% agarose gel electrophoresis [Thermo Fisher Scientific; Victoria, AUS].
Sensitivity of the optimised qPCR assay
The sensitivity threshold of the qPCR assay was evaluated using a 10-fold serial dilution of S. aureus AU-19 with a starting gDNA concentration of 10ng/µl determined by a Nanodrop Spectrophotometer [Thermo Fisher Scientific; Victoria, AUS]. The qPCR assay was performed with the Takara SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) Detection Kit [Scientifix; Victoria, AUS] using the Real-Time CFX Connect PCR System [BioRad; California, USA]. All reactions were performed in triplicate and included both positive and negative controls. The qPCR assay was performed as per the manufacturer’s instructions32 using primers specific to the nuc gene24.
Clinical PICC sample gDNA extraction
Following the semi-quantitative roll-plate culture, clinical PICC samples were individually suspended overnight in 200µl of lysis buffer which contained 20mg/mL of lysozyme, 20mm Tris-HCl (pH 8.0), 2mm EDTA, 1.2% Triton, and Proteinase K at 37˚C. Bacterial gDNA was extracted from each PICC using the PowerSoil DNA Isolation Kit [Mo Bio; California, USA].
Validating the sensitivity of the qPCR assay using clinical PICC samples
The qPCR assay was performed directly on extracted bacterial gDNA from each clinical PICC sample. The qPCR assay was performed with the Takara SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) Detection Kit [Scientifix; Victoria, AUS] using the Real-Time CFX Connect PCR System [BioRad; California, USA]. All reactions were performed in triplicate and included both positive and negative controls according to the manufacturer’s instructions32, using primers specific to the nuc gene24.
RESULTS
Conventional PCR: amplification of the nuc gene
The nuc gene primers24 successfully amplified S. aureus AU-19 as the target amplicon with the PCR product amplified as a 270-bp fragment visualised using 2% agarose gel electrophoresis [Thermo Fisher Scientific; Victoria, AUS] (Figure 1).
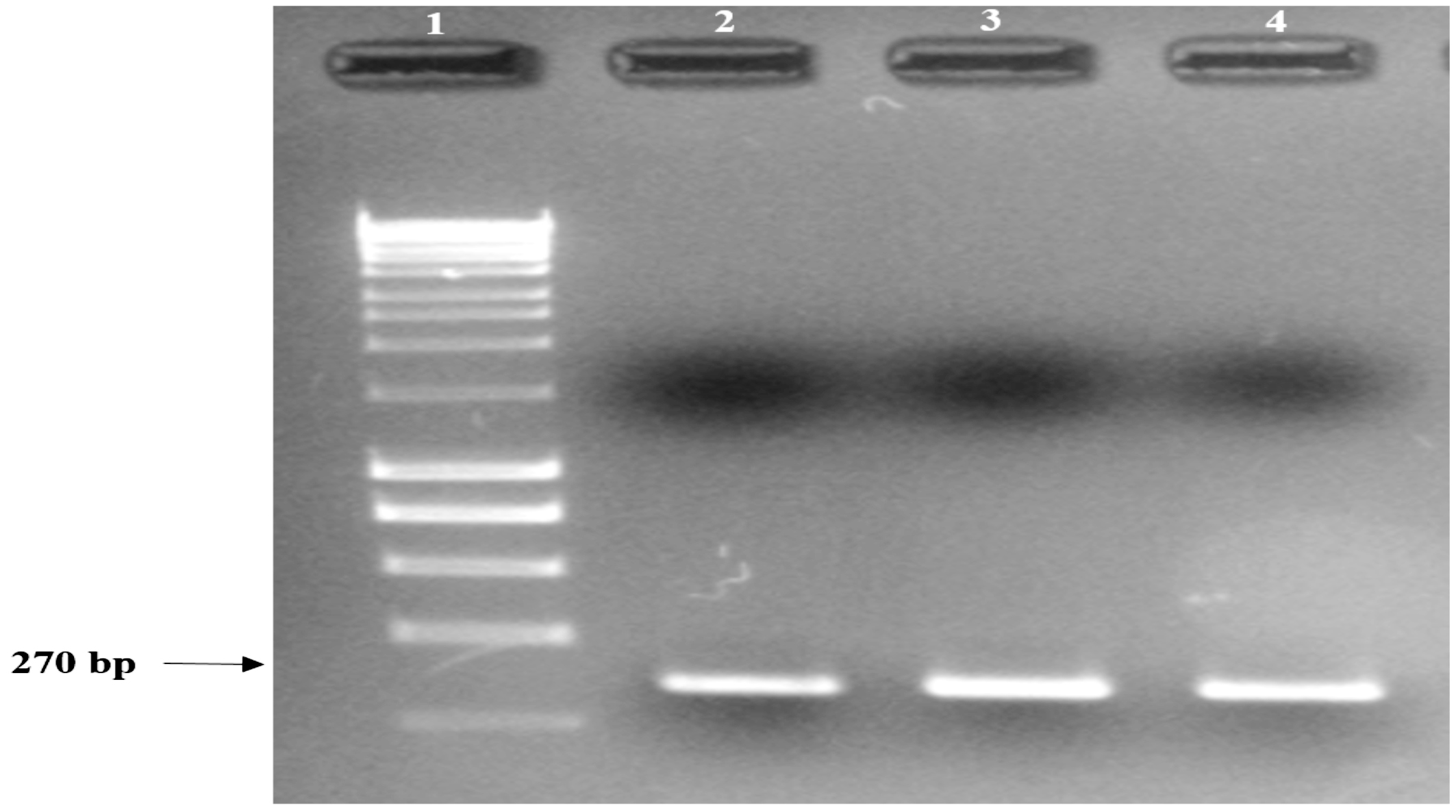
Figure 1. S. aureus AU-19 conventional PCR product showing amplification of the expected size fragment obtained with nuc1 – nuc2 primers, analysed under UV light using 2% agarose gel.
Lane 1: 1 Kbp molecular weight marker;
Lane 2: S. aureus AU-19 PCR product replicate 1;
Lane 3: S. aureus AU-19 PCR product replicate 2;
Lane 4: S. aureus AU-19 PCR product replicate 3.
Sensitivity of the optimised qPCR assay
The qPCR assay detection limit was 102 CFU/mL-1 (Cq 33.63 ± 0.58) using a 10-fold serial dilution of S. aureus AU-19 gDNA. The Cq values of the 10-fold serial dilution (106 to 101 CFU/mL-1) ranged from Cq 15.01 ± 0.13 to Cq 37.46 ± 1.18, respectively. Non-specific amplification was observed after 35 cycles. The Real-Time CFX Connect PCR System [BioRad; California, USA] analysis software indicated an efficiency of 98.8%, correlation coefficient (R2)=0.974 and slope of the graph (y)=-3.352 (Figure 2).
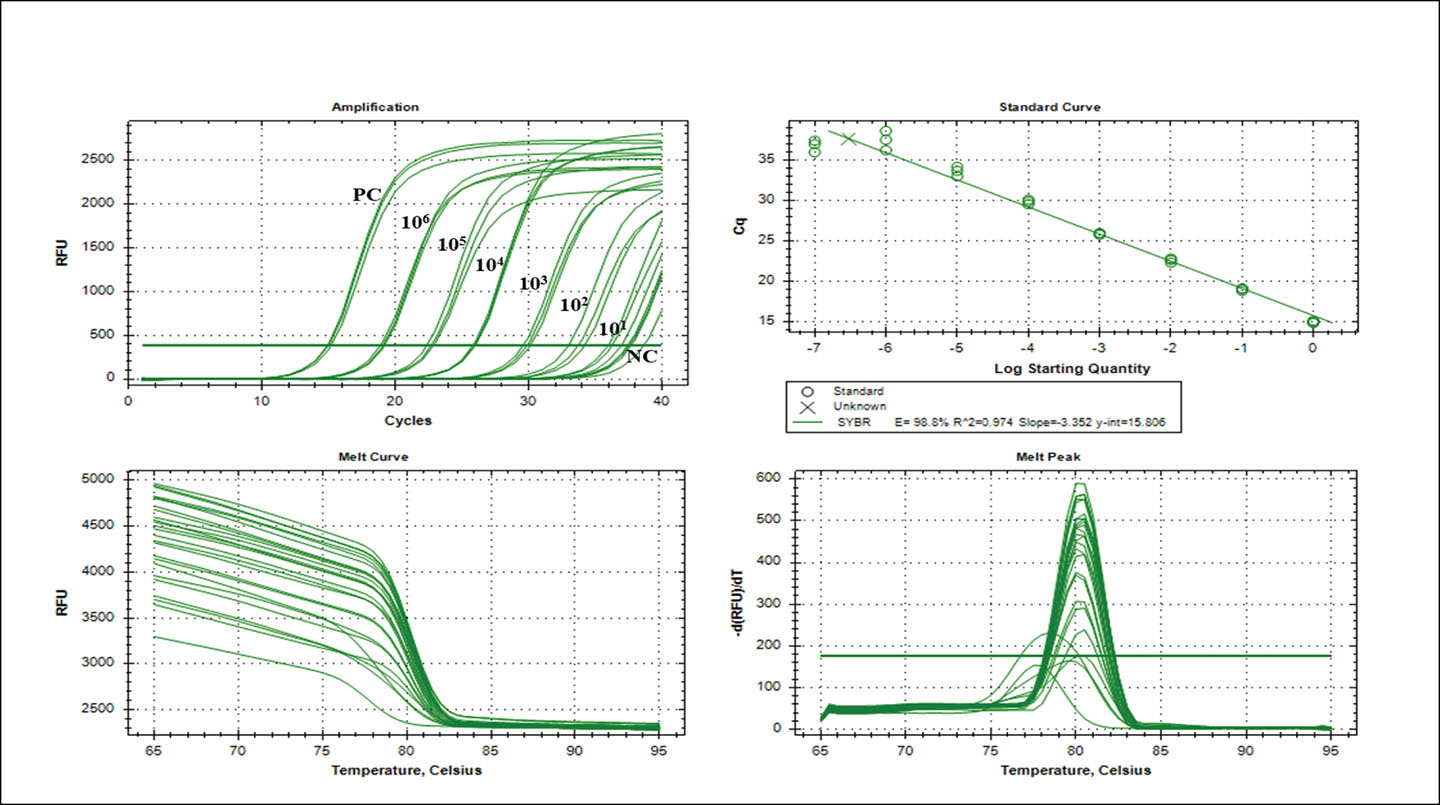
Figure 2. Real-time qPCR assay. Amplification of the nuc gene (106–101 nuc gene copy) using gDNA isolated from an overnight culture of S. aureus AU-19. The standard curve plotted using the log starting quantity against the mean Cq. Data from the straight lines calculated by linear regression yielded an efficiency of 98.8%, correlation coefficient (R2)=0.974 and slope of the graph (y)=-3.352. Error bars indicate the standard deviations based on three replications.
Validation of the qPCR assay using clinical PICC samples
The qPCR assay was validated using a total of 41 clinical PICC samples. One PICC produced positive amplification (Cq 29.82 ± 0.18) equivalent to 103 CFU/mL-1 using the nuc gene primers22. Non-specific amplification was observed after 35 cycles. The Real-Time CFX Connect PCR System [BioRad; California, USA] analysis software indicated an efficiency of 96.7%, correlation coefficient (R2)=0.989 and slope of the graph (y)=-3.403 (Figure 3). The results of the qPCR assay correlated (100%) with the results from the semi-quantitative roll-plate culture method (103 CFU/mL-1).
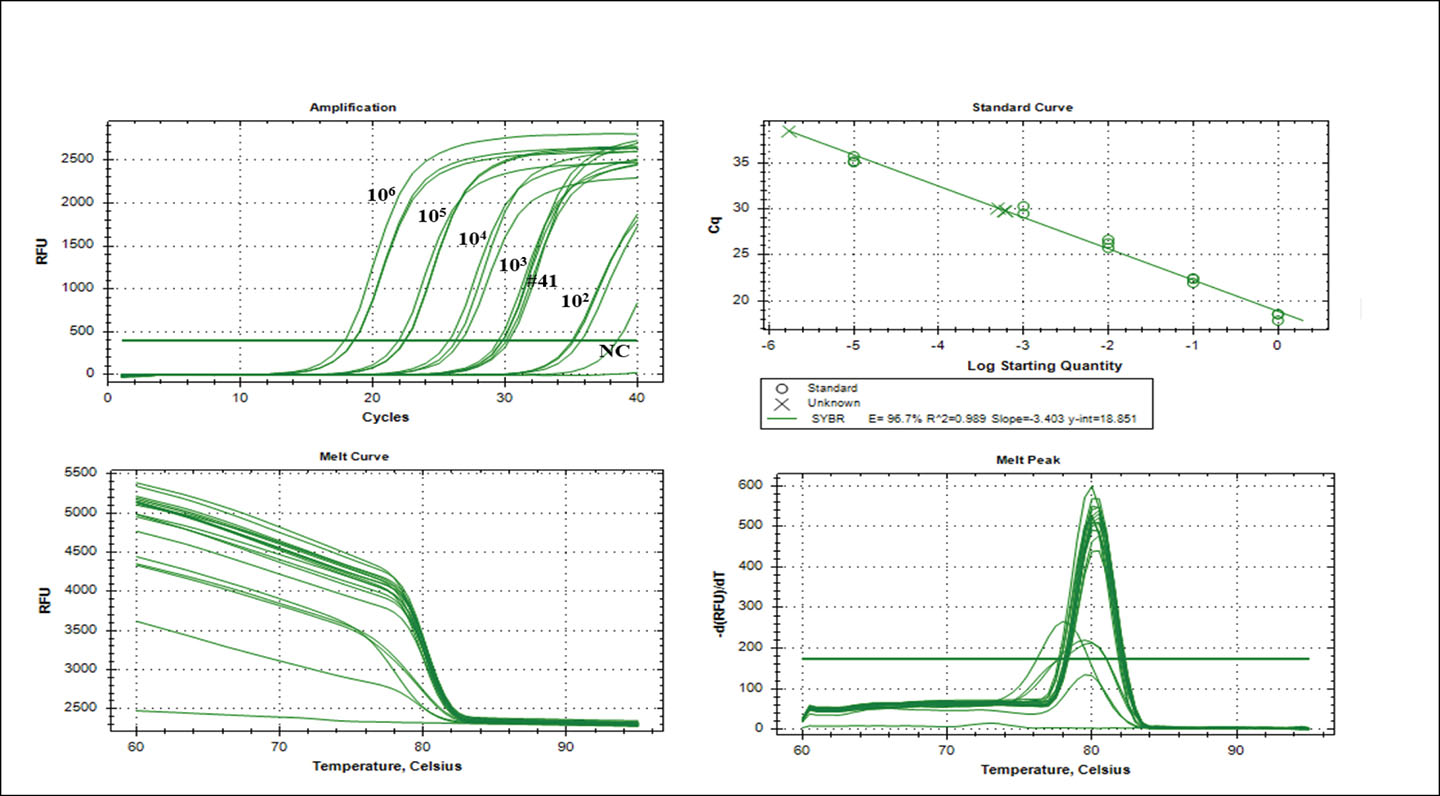
Figure 3. Real-time qPCR assay. Amplification of the nuc gene (106–102 nuc gene copy) using gDNA isolated from an overnight culture of S. aureus AU-19. One clinical PICC sample was amplified at Cq 29.82 ± 0.18. The standard curve plotted using the log starting quantity against the mean Cq. The straight lines calculated by linear regression and the data yielded an efficiency of 96.7%, correlation coefficient (R2)=0.989 and slope of the graph (y)=-3.403. Error bars indicate the standard deviations based on three replications.
DISCUSSION
Molecular diagnostic methods including qPCR are becoming widely accepted as methods of choice for accurate and rapid detection of bacteria in clinical samples33. The qPCR assay developed in this study has proved to be a time- and cost-effective high-throughput method. It can be performed directly on isolated bacterial gDNA with a processing time of less than 2 hours after gDNA extraction22. Another advantage of this qPCR assay is that amplification reactions are analysed in a closed system, eliminating the need for post-amplification manipulation and therefore reducing opportunities for contamination27.
The nuc gene primers used in this study were selected based on their specificity for the S. aureus genome34,35. The 270-bp fragment of the nuc gene was successfully amplified using conventional PCR and optimised for the qPCR assay. Brakstad and colleagues (1992) originally proved the suitability of the nuc gene for the detection of S. aureus and the results of this study agree with their claims24.
The sensitivity threshold of this qPCR assay, with the reference strain S. aureus AU-19, was equivalent to 102 CFU/mL-1 (Cq 33.63 ± 0.58) after overnight enrichment culture. Non-specific amplification was observed after 35 cycles. The occurrence of non-specific amplification, such as primer dimer, can be overcome with further optimisation of the qPCR assay or the incorporation of a fluorescently labelled oligonucleotide probe36. Hoegh and colleagues (2014) reported that using qPCR nuc gene-specific primers alone for detection and identification of S. aureus may result in misidentification of certain strains of S. aureus and that additional species-specific targets should be considered to increase assay efficiency37. Numerous qPCR-based assays targeting the nuc gene alone or in combination with the mecA gene have been described for the rapid screening or identification of S. aureus, including both methicillin-sensitive and methicillin-resistant strains of S. aureus25,27,38.
In total, 41 clinical PICC samples were used in this study to validate the sensitivity of the qPCR assay by determining the detection level of S. aureus. One of the 41 clinical PICC samples was found to be colonised (103 CFU/mL-1) by both the qPCR assay and the semi-quantitative roll-plate culture method. On further analysis, the patient was diagnosed with CLABSI and isolates from both the PICC and patient’s bloodstream were identified as S. aureus. Therefore, this qPCR assay proved to be an accurate and rapid detection method for S. aureus PICC colonisation.
Although this study had limitations, including a small sample size, and only one patient with confirmed S. aureus PICC colonisation and subsequent CLABSI, it does present a good model and demonstrates promise as a suitable method for the detection of S. aureus PICC colonisation.
The qPCR assay, using primers specific to the nuc gene used in this study, accurately and rapidly detected S. aureus PICC colonisation. With further optimisation and validation with a larger sample size, this qPCR assay has the potential to be used as a diagnostic method in a clinical setting.
Acknowledgement
The authors thank all clinical staff within the Vascular Assessment and Management Service in Queensland Children’s Hospital and AVATAR group for their efforts in recruiting patients, data collecting, and ensuring protocol adherence.
Author(s)
*Maddie Higgins1; Jeremy Brownlie1; Li Zhang2,3; Rebecca Ford1
1School of Environment and Science, Griffith University, Brisbane, Australia
2School of Dentistry and Oral Health, Gold Coast Campus, Griffith University, Gold Coast, Australia
3Alliance for Vascular Access Teaching and Research (AVATAR), Menzies Health Institute Queensland, Griffith University, Brisbane, Australia
*Corresponding author
Maddie Higgins, School of Environment and Science, Griffith University, Brisbane, Australia
Email maddiejhiggins@bigpond.com
References
1. Bowen A, Carapetis J. Advances in the diagnosis and management of central venous access device infections in children. Adv Exp Med Biol 2011;697:91–106.
2. Safdar N, Maki DG. Risk of catheter-related bloodstream infection with peripherally inserted central venous catheters used in hospitalized patients. Chest 2005 Aug;28(2):289-95.
3. Timsit JF. Diagnosis and prevention of catheter-related infections. Curr Opin Crit Care 2007;13(5):563–71.
4. Zhang L, Gowardman JR, Rickard C. Impact of microbial attachment on intravascular catheter-related infections. Int J Antimicrob Agents 2011;1:9–15.
5. Blot SI, Depuydt P, Annemans L, Benoit D, Hoste E, Jan JDW, et al. Clinical and economic outcomes in critically ill patients with nosocomial catheter-related bloodstream infections. Clin Infec Dis 2005;41(11):1591–8.
6. Warren KD, Quadir WW, Hollenbeak SC, Elward MA, Cox JM, Fraser JV. Attributable cost of catheter-associated bloodstream infections among intensive care patients in a nonteaching hospital. Crit Care Med 2006;34(8):2084–9.
7. Mermel LA, Allon M, Bouza E, Craven DE, Flynn P, O’Grady NP, et al. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin Infec Dis 2009;49(1):1–45.
8. O’Grady NP, Alexander M, Burns LA, Dellinger EP, Garland J, Heard SO, et al. Guidelines for the prevention of intravascular catheter-related infections. Clin Infec Dis 2011;52(9):e162–e93.
9. Maki DG, Kluger DM, Crnich CJ. The risk of bloodstream infection in adults with different intravascular devices: a systematic review of 200 published prospective studies. Proceedings of the Mayo Clinic 2006;81(9):1159–71.
10. Goudie A, Dynan L, Brady PW, Rettiganti M. Attributable cost and length of stay for central line-associated bloodstream infections. Pediatrics 2014;133(6):e1525–e32.
11. Stevens V, Geiger K, Concannon C, Nelson RE, Brown J, Dumyati G. Inpatient costs, mortality and 30-day re-admission in patients with central-line-associated bloodstream infections. Clin Microbiol Infec 2014;20(5):O318–O24.
12. Gaynes R, Edwards JR, The National Nosocomial Infections Surveillance S. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis 2005;41(6):848–54.
13. Collignon P, Cruickshank M, Dreimanis D. Staphylococcus aureus bloodstream infections: an important indicator for infection control. Chapter 2: Bloodstream infections an abridged version. Healthcare Infection 2009;14(4):165–71.
14. Kornbau C, Lee K, Hughes G, Firstenberg M. Central line complications. Int J Crit Ill Inj Sci 2015;5:170.
15. Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis (major article). Clin Infect Dis 2003;36(1):53.
16. Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol 2008;8(6):747–63.
17. Alarcón B, Vicedo B, Aznar R. PCR-based procedures for detection and quantification of Staphylococcus aureus and their application in food. J Appl Microbiol 2006;100(2):352–64.
18. Beutz M, Sherman G, Mayfield J, Fraser VJ, Kollef MH. Clinical utility of blood cultures drawn from central vein catheters and peripheral venipuncture in critically ill medical patients. Chest 2003;123(3):854.
19. Grace C, Lieberman J, Pierce K, Littenberg B. Usefulness of blood culture for hospitalized patients who are receiving antibiotic therapy. Clin Infec Dis 2001;32(11):1651–5.
20. Munson EL, Diekema DJ, Beekmann SE, Chapin KC, Doern GV. Detection and treatment of bloodstream infection: laboratory reporting and antimicrobial management. J Clin Microbiol 2003;41(1):495.
21. Peters RP, Mohammadi T, Vandenbroucke-Grauls CM, Danner SA, van Agtmael MA, Savelkoul PH. Detection of bacterial DNA in blood samples from febrile patients: underestimated infection or emerging contamination? FEMS Immunol Med Microbiol 2004;42(2):249–53.
22. Remco PHP, Michiel AvA, Gierveld S, Danner SA, Groeneveld ABJ, Christina MJEV-G, et al. Quantitative detection of Staphylococcus aureus and Enterococcus faecalis DNA in blood to diagnose bacteremia in patients in the intensive care unit. J Clin Microbiol 2007;45(11):3641–6.
23. Wang H-Y, Kim S, Kim H, Kim J, Kim Y, Park S-D, et al. Real-time PCR TaqMan assay for rapid screening of bloodstream infection. Ann Clin Microbiol Antimicrob 2014;13(1):3.
24. Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol 1992;30(7):1654–60.
25. Thomas LC, Gidding HF, Ginn AN, Olma T, Iredell J. Development of a real-time Staphylococcus aureus and MRSA (SAM-) PCR for routine blood culture. J Microbiol Meth 2007;68(2):296–302.
26. Cattoir V, Merabet L, Djibo N, Rioux C, Legrand P, Girou E, et al. Clinical impact of a real-time PCR assay for rapid identification of Staphylococcus aureus and determination of methicillin resistance from positive blood cultures. Clin Microbiol Infec 2011;17(3):425–31.
27. Costa A-M, Kay I, Palladino S. Rapid detection of mecA and nuc genes in staphylococci by real-time multiplex polymerase chain reaction. Diagn Micr Infec Dis 2005;51(1):13–7.
28. Children’s Health Services. CVAD insertion and management guideline. Brisbane: Queensland Government; 2014.
29. Maki DG, Weise CE, Sarafin HW. A semiquantitative culture method for identifying intravenous-catheter-related infection. N Engl J Med 1977;296(23):1305.
30. Centers for Disease Control and Prevention. National healthcare safety network device associated module: CLABSI. Atlanta: USA Government; 2014. p. 1–9.
31. Bio-Rad. MyCycler thermal cycler instruction manual. California: Bio-Rad Laboratories, Inc; no date.
32. Bio-Rad. Real-time PCR application guide. California: Bio-Rad Laboratories, Inc; 2006.
33. Paolucci M, Landini MP, Sambri V. Conventional and molecular techniques for the early diagnosis of bacteraemia. Int J Antimicrob Agents 2010;36(2):S6–S16.
34. Maes N, Magdalena J, Rottiers S, De Gheldre Y, Struelens MJ. Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative staphylococci and determine methicillin resistance from blood cultures. J Clin Microbiol 2002;40(4):1514.
35. Louie L, Goodfellow J, Mathieu P, Glatt A, Louie M, Simor AE. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J Clin Microbiol 2002;40(8):2786.
36. Hein I, Lehner A, Rieck P, Klein K, Brandl E, Wagner M. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl Environ Microb 2001;67(7):3122–6.
37. Hoegh SV, Skov MN, Boye K, Worning P, Jensen TG, Kemp M. Variations in the Staphylococcus aureus-specific nuc gene can potentially lead to misidentification of methicillin-susceptible and -resistant S. aureus. J Med Microbiol 2014;63(Pt 7):1020–2.
38. Elsayed S, Chow B, Hamilton N, Gregson D. Development and validation of a molecular beacon probe-based real-time polymerase chain reaction assay for rapid detection of methicillin resistance in Staphylococcus aureus. Arch Pathol Lab Med 2003;127(7):845–9.
